CANCER EPIGENOME CENTER
A DIFFERENT APPROACH TO FIGHTING CANCER
-A new center aims to combat cancer by rewriting the epigenetic modifications harbored by tumor cells.
Research Keywords: Cancer, Epigenome, Inhibitor
 iStock.com/vitanovski
iStock.com/vitanovskiCancer is thought of as a genetic disease, with mutations to a cell’s DNA causing a normal cell to give birth to a malignant tumor. Yet epigenetic alterations, which leave the DNA sequence intact, can be just as important in a cell’s transformation to cancer. Even though epigenetic regulation through mechanisms such as methyl tags and histone modifications does not alter the DNA sequence, it can chemically modify the genome to signal which genes should be turned on and off. When tumor-suppressor genes are silenced or oncogenes are activated in this way, many different forms of cancer can result.
Fortunately for medicine, epigenetic alterations are not permanent. They can be undone to prevent or reverse cancer, leaving a cell with its entire complement of normal DNA intact.
This is exactly what researchers at the Cancer Epigenome Center are striving to do. Led by Atsushi Kaneda, the center is seeking to understand the various epigenetic drivers that contribute to different types of cancer and then develop new drugs that overturn these effects.
“We will explain key epigenomic aberrations and their molecular causes,” says Kaneda. “And then, taking advantage of small molecules that bind to DNA in a sequence-specific manner, we will develop drugs that can rewrite the accumulated epigenomic aberrations or prevent them accumulating in targeted genomic regions.”
UNRAVELING MOLECULAR MECHANISMS
Kaneda points to ongoing investigations of gastric cancer as an example of this strategy in action. Patients with this cancer exhibit different levels of DNA methylation in their tumor cells, depending on the pathogen responsible for the disease. The cancer-causing bacterium Helicobacter pylori is one source of irregular methylation, infection with Epstein Barr virus is another. Both seem to have profound epigenetic impacts on stomach cells, leading to the silencing of many tumor suppressors.
Kaneda and his colleagues are using clinical samples and cell experiments to unravel the molecular mechanisms that induce hyperactive methylation patterns following infection with these pathogenic agents. They ultimately hope to stop the process in its tracks.
Similar projects are ongoing to combat colorectal cancers — which Chiba researchers have shown can be categorized into three types, depending on methylation levels — as well as blood cancers and other types of tumor.With an eye to developing novel therapeutics, chemists at the Cancer Epigenome Center are working on new methods to efficiently synthesize molecules that can act on epigenetic aberrations. One such group of molecules is the pyrrole-imidazole polyamides. These molecules can bind to DNA at specific sites in the genome, and when coupled to an epigenetic inhibitor they can prevent methylation-based gene silencing. Kaneda hopes to deploy the molecules to edit the epigenome at targeted regions to regulate tumor formation.
The Cancer Epigenome Center includes researchers from a wide range of backgrounds and specialties. This multidisciplinary team approach, says Kaneda, is essential for turning the center’s discoveries into clinically meaningful treatments and diagnostic tools. “We will help determine the mechanisms of cancers and develop new cancer drugs through collaborating with experts in medicine, physics and pharmacy,” he says.
(CHIBA RESEARCH 2020)Members
Principal Investigator
| Name | Title, Affiliation | Research Themes |
|---|---|---|
| KANEDA Atsushi | Professor, Graduate School of Medicine | Cancer Epigenomics |
Co-Investigatior
| Name | Title, Affiliation | Research Themes |
|---|---|---|
| NEMOTO Tetsuhiro | Professor, Graduate School of Pharmaceutical Science | Pharmaceutical Chemistry |
| URA Kiyoe | Professor, Graduate School of Science | Molecular Biology Genetics |
| MATSUBARA Hisahiro | Professor, Graduate School of Medicine | Digestive Cancer |
| YOSHINO Ichiro | Professor, Graduate School of Medicine | Lung Tumor |
| HANAOKA Hideki | Professor, Chiba University Hospital | Head and Neck Tumor |
| MANABE Ichiro | Professor, Graduate School of Medicine | Lymphedema |
| TANAKA Tomoaki | Professor, Graduate School of Medicine | Endocrinology, Cell Senescence |
| ICHIKAWA Tomohiko | Professor, Graduate School of Medicine | Cancer Urinary |
| MATSUE Hiroyuki | Professor, Graduate School of Medicine | Ecpyma |
| SAKAIDA Emiko | Medical Professor, Chiba University Hospital | Blood Tumor |
| HANAZAWA Toyoyuki | Professor, Chiba University Hospital | Head and Neck Tumor |
| SHINAGAWA Yoko | Associate Professor, Academic Research & Innovation Management Organization | Intellectual Property Strategics |
Research report(2016〜2018)
Cancer arises through accumulation of epigenetic and genetic alterations, and therefore is stratified into several molecular subtypes using comprehensive epigenomic and genomic information. Elucidation of tumorigenic mechanism for each subtype and establishment of specific target therapy are pressing issues for personalized/stratified medicine against cancer, the leading cause of death in our country.
In this project, we established cancer epigenome center to elucidate epigenomic aberrations in tumorigenesis and develop small molecules to target pathological aberrations, based on the research network we had formed in Start-up COE Program of Chiba University since 2013. The center gathered clinical departments to supply clinical cancer specimens, basic biology laboratories to condut detailed epigenomic analyses using in vitro and/or in vivo cancer models, and pharmaceutical chemistry laboratories to synthesize small molecules to to edit altered epigenome.
We conducted stratification of caner into distinct molecular subtypes through epigenomic analyses in many types of cancer, including gastric cancer, colorectal cancer, lung cancer, hematopoietic malignancies, etc. Gastric cancer, for example, was stratified into several DNA methylation epigenotypes depend on pathogens. We have shown using our in vitro infection system (Fig 1) that Epstein Barr virus (EBV) infection is the cause of extensive hypermethylation observed in EBV+ gastric cancer, and that repression of a demethylating enzyme by viral transcripts or upregulated human miRNAs is a cause of DNA methylation induction. Other epigenomic modifiers were also involved to alter epigenomic status in promoter regions, enhancer regions, etc, during EBV infection. Histone activation marks were acquired at enhancers of genes for cellular proliferation and undifferentiated state, and lost at enhancers for growth suppression and epithelial differentiation, leading to alteration of cell fate in gastric epithelial cells from differentiated/unproliferative state to undifferentiated/proliferative state.
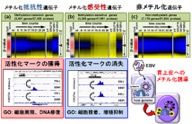 Figure 1. Epigenomic analyses using in vitro EBV infection system. We elucidated dynamic epigenomic alterations including DNA methylation and histone active marks in gastric epithelial cells during EBV infection.
Figure 1. Epigenomic analyses using in vitro EBV infection system. We elucidated dynamic epigenomic alterations including DNA methylation and histone active marks in gastric epithelial cells during EBV infection.
Using in vitro systems and clinical tissue samples, we elucidated key epigenomic aberrations and their molecular causes. Taking advantage by utilizing small molecules that bind to DNA by sequence-specific manner, we developed small molecules that could rewrite the accumulated epigenomic aberrations or prevent their accumulations in targeted genomic regions. Pyrrole (P) Imidazole (I) polyamide is a class of small molecules that bind to the minor groove of DNA. PI polyamides are hydrophobic, cell-permeable and intravenously injectable small molecules with low toxicity. We established PI polyamide that recognizes gene promoter sequence and inhibits induction of DNA methylation. We also conjugated small molecule epigenetic inhibitor to PI polyamide, which was delivered to the targeted genomic regions recongnized by the conjugated PI polyamide. We successfully established conjugates to alter histone modifications in specific genomic regions by sequence-specific manner, and filed patents for these techniques to edit epigenome and novel methods to efficiently synthesize PI polyamides.
